
Jupyter Enterprise Gateway
In this blog post I would like to share with you my experience with Jupyter Enterprise Gateway, a headless web server with a pluggable framework for anyone supporting multiple Jupiter Notebook users in a managed-cluster environment. I am part of a DevOps team that manages the infrastructure for several data scientist teams, and we needed a solution to divide our pool of crunching resources amongst them. To do this, we chose Jupiter Enterprise Gateway.
What is project Jupyter?
The Jupyter Enterprise Gateway is part of the
Jupyter project, an non-profit that aims to develop open source software, open standards and other services that enable interactive data science. This project is probably most known amongst data scientists for their Jupyter Notebook product.
What are Jupyter Notebooks?
To explain Jupyter Notebooks we can go to the
docs of the Jupyter eco system and we will read the following.
Jupyter Notebooks are structured data that represent your code, metadata, content, and outputs. When saved to disk, the notebook uses the extension .ipynb, and uses a JSON structure.
This technology is used by data scientists mainly to do development work. You can easily test code or cells, refactor them and try again. While there are obvious differences, you could compare Jupyter Notebooks to an IDE used by software developers.
What is Jupyter Enterprise Gateway?
From a technical perspective, Jupyter Enterprise Gateway is a system that gives you the ability to launch kernels on behalf of remote notebooks. This way you can manage resources better, as the system is no longer the single location for kernel activity. It essentially exposes a Kernel as a Service model.
By default, the Jupyter framework runs kernels locally. When using an orchestration system like Kubernetes, Jupyter Enterprise Gateway distributes kernels across the cluster, dramatically increasing the number of simultaneously active kernels while leveraging the available compute resources.
Why we needed Jupyter Enterprise Gateway
The main reason we needed Jupyter Enterprise Gateway is because we have a Kubernetes cluster that is shared amongst multiple teams of data scientists. We needed a solution to efficiently expose the resources of our cluster to different users and groups.
How are we going to expose these resources in an efficient way to different users or groups?
With Jupyter Enterprise Gateway we were able to do this. After we deployed the application in our cluster, we could now provide our teams of data scientists with simultaneous kernels.
Which problem does Jupyter Enterprise Gateway solve?
With Jupyter Enterprise Gateway we accomplish the fact that high cpu, memory or gpu workloads can be offloaded to a our Kubernetes cluster instead of your (the data scientist’s) local machine. We now have one centralized source of resources.
End users will use a Jupyter command to connect to the gateway, this will create a Jupyter server on their own localhost. The Jupyter server on their localhost will connect to the Jupyter enterprise gateway running in the cluster.
Kernels in Kubernetes?
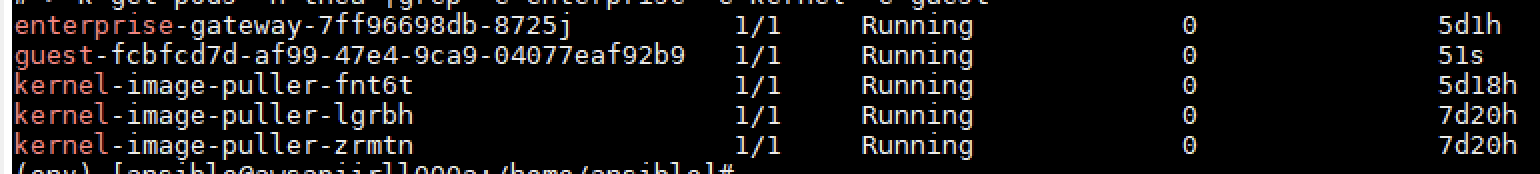
Yes. All of the “standard” environments are available from the get go ; R, Python, Scala,… And all kernels that the end user create are represented by pods in the Kubernetes cluster running the Jupyter Enterprise Gateway.
To get the images to the nodes Jupyter Enterprise Gateway also provides kernel image pullers to handle the image pulling for you. From the moment a kernel gets scheduled on a node the kernel image puller will start the pulling of the given image that is needed in the kernel.
Custom kernels?
Yep. Every docker image that we use can be edited to be a Jupyter enterprise gateway kernel. We just need to install a couple of extra python packages in our image which is pretty straightforward. However, the extra catch is when we tried running Conda environments in our docker image. This was a little bit more tricky, which I will explain below.
All kernel images are defined by three files.
- kernel.json
- kernel-pod.yaml.j2
- launch-kubernetes.py
More info on how to customize kernels can be found here.
The tricky part for Conda environments is this file. The bootstrap-kernel.sh file will take care of -exactly- bootstrapping a new kernel. This is done using a python command. To enter a kernel in an activated Conda environment, we needed to edit this python reference to the reference from our Conda environment.
On line 26 you can find the following command, for Conda alter the python command to /opt/conda/envs/_envname_/bin/python.
python ${KERNEL_LAUNCHERS_DIR}/python/scripts/launch_ipykernel.py --kernel-id ${KERNEL_ID} \
This bootstrap-kernel.sh file is needed IN the image. For this reason we incorporated this step in the image building process.
Everything deployed, what did we win with this solution?
In the end Jupyter Enterprise Gateway is a layer on top of our cluster to make it easier for end users to create notebooks on demand that use the resources of the cluster. So from end user perspective, the data scientists are just creating a new notebook server on their localhost but with the advantage of having a Kubernetes cluster backing up the kernels with a lot more resources than they would ever be able to get on their local machine.
We are using the Jupyter Enterprise Gateway as a development tool. In the end after development we will still create longer running Kubernetes jobs to do the actual training.
I hope that the experience I shared will help to make your journey into Jupyter Enterprise Gateway a little bit easier.
Jupyter Enterprise Gateway Github



